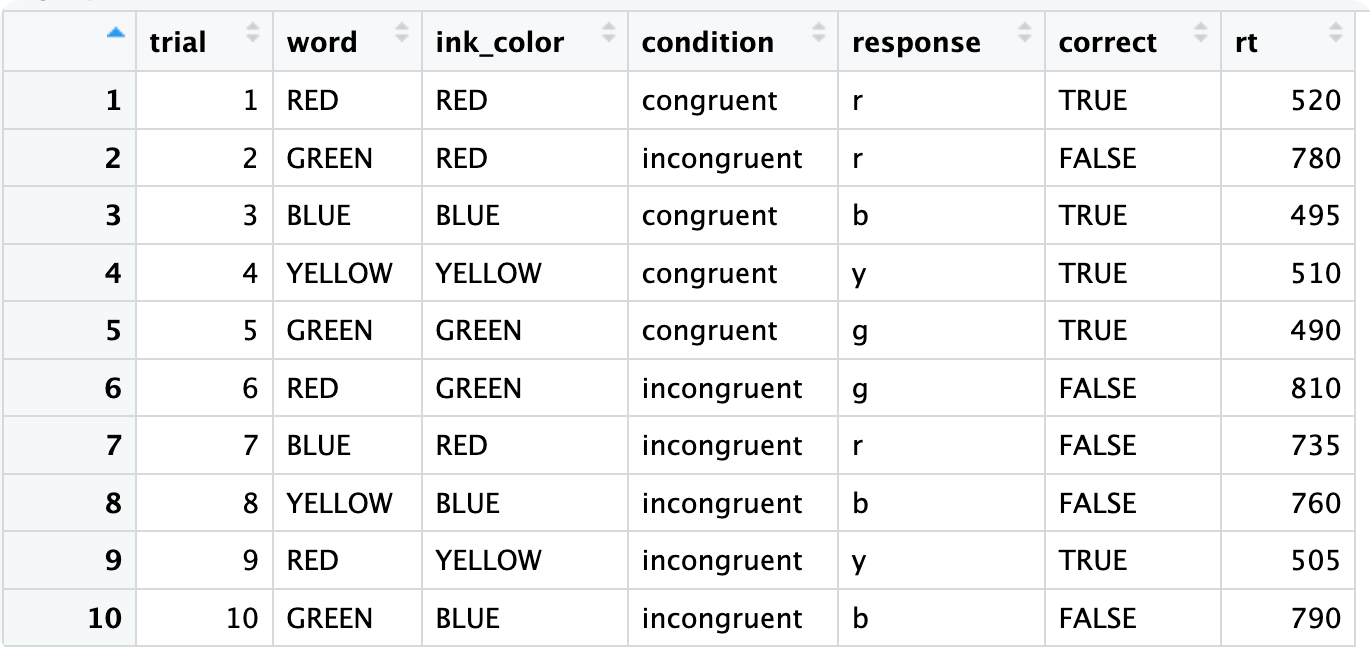
Let’s explore how R utilizes loops and vectorized operations — two different approaches to performing repetitive tasks. We’ll use a small Stroop Task dataset as our example:
stroop_data <- data.frame(
trial = 1:10,
word = c("RED", "GREEN", "BLUE", "YELLOW", "GREEN", "RED", "BLUE", "YELLOW", "RED", "GREEN"),
ink_color = c("RED", "RED", "BLUE", "YELLOW", "GREEN", "GREEN", "RED", "BLUE", "YELLOW", "BLUE"),
condition = c("congruent", "incongruent", "congruent", "congruent",
"congruent", "incongruent", "incongruent", "incongruent",
"incongruent", "incongruent"),
response = c("r", "r", "b", "y", "g", "g", "r", "b", "y", "b"),
correct = c(TRUE, FALSE, TRUE, TRUE, TRUE, FALSE, FALSE, FALSE, TRUE, FALSE),
rt = c(520, 780, 495, 510, 490, 810, 735, 760, 505, 790) # reaction times (ms)
)
Most programming languages, including R, provide loops which are programming structures that let you repeat a block of code multiple times, either for a fixed number of iterations or until a condition is met.
Here’s a simple example of a for loop in R:
for (i in 1:5) {
print(paste("This is loop number", i))
}
We can use loops to perform operations on data frame rows. For example, the following code adds a new variable rt_category to our Stroop dataset, categorizing each participant’s response time as fast (≤ 500 ms) or slow(everything else):
# Create a new (empty) column
stroop_data$rt_category <- NA
# Use a for loop to classify each subject
for (i in 1:nrow(stroop_data)) {
if (stroop_data[i, "rt"] <= 500) {
stroop_data[i, "rt_category"] <- "fast"
} else {
stroop_data[i, "rt_category"] <- "slow"
}
}
The above approach works, but it doesn’t take advantage of one of R’s most powerful features: vectorization.
Many R functions are vectorized, meaning they can operate on an entire vector or column at once rather than looping through individual elements.
Here’s how to rewrite the same operation using R’s vectorized ifelse() function:
stroop_data$rt_category <- ifelse(stroop_data$rt < 500, "fast", "slow")
In this version, ifelse() checks the entire stroop_data$rt column in a single step.
For each element:
stroop_data$rt < 500 is TRUE, then "fast" is assigned.FALS then "slow" is assigned.The result is a new vector of the same length as the input column- no explicit looping required.
Vector operations are usually preferred in R for two main reasons:
Simpler and more expressive code - They’re shorter, easier to read, and less error-prone. Less code = fewer mistakes.
Speed and efficiency - Vectorized functions are typically 10–100× faster than explicit loops because they’re optimized internally in compiled code. They perform calculations on whole vectors at once, rather than one element at a time.
While vectorized code is ideal for data manipulation and calculations, loops are still useful in several situations:
For example, here’s a simple loop that reads all CSV files in a folder and prints a summary of each one:
for (file in list.files("data", full.names = TRUE)) {
df <- read.csv(file)
print(summary(df))
}
You could achieve the same result using the vectorized lapply() function:
files <- list.files("data", full.names = TRUE)
lapply(files, function(file) {
df <- read.csv(file)
summary(df)
})
In this case, both approaches work but a loop may feel more natural or readable.
Loops are especially useful when generating or storing results step by step. For example, the code below simulates sampling variability by computing the mean of 10 random uniform numbers across 100 samples:
results <- numeric(100)
for (i in 1:100) {
results[i] <- mean(runif(10))
}
Each iteration draws 10 random numbers, computes their mean, and stores it in the vector results.
For a fun example of simulations and loops in R, check out this blog post: What is the probability that two persons have the same initials?
No subscriptions, no auto-renewals.
Just a simple one-time payment that helps support my free, to-the-point videos without sponsered ads.
Unlocking gets you access to the notes for this video plus all 200+ guides on this site.
Your support is appreciated. Thank you!